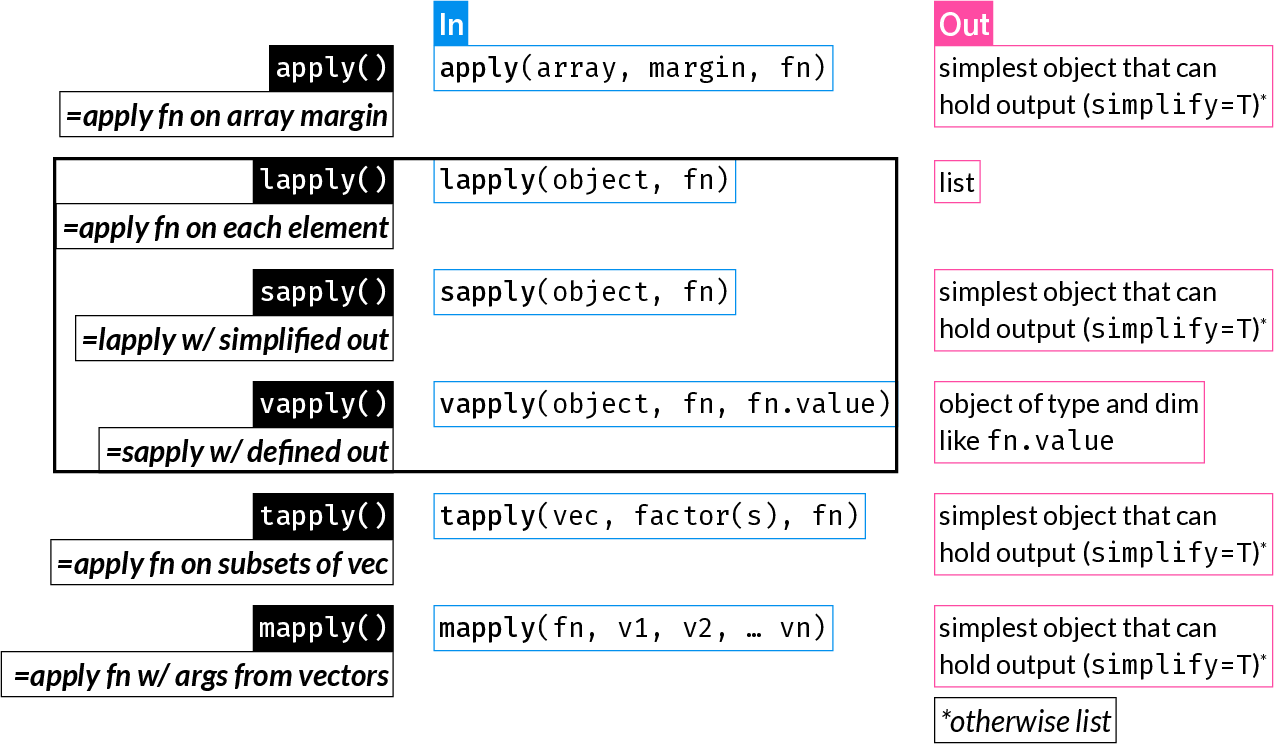
24 The Apply Family
The apply functions are some of the most widely used R functions. They replace longer expressions created with a for loop, for example.
They can result in more compact and readable code.
| Function | Description |
|---|---|
apply() |
Apply function over array margins (i.e. over one or more dimensions) |
lapply() |
Return a list where each element is the result of applying a function to each element of the input |
sapply() |
Same as lapply(), but returns the simplest possible R object (instead of always returning a list) |
vapply() |
Same as sapply(), but with a pre-specified return type: this is safer and may also be faster |
tapply() |
Apply a function to elements of groups defined by a factor |
mapply() |
Multivariate sapply(): Apply a function using the 1st elements of the inputs vectors, then using the 2nd, 3rd, etc. |
24.1 apply()
apply() applies a function over one or more dimensions of an array of 2 dimensions or more (this includes matrices) or a data frame:
apply(array, MARGIN, FUN)
MARGIN can be an integer vector or character indicating the dimensions over which ‘FUN’ will be applied.
By convention, rows come first (just like in indexing), therefore:
-
MARGIN = 1: apply function on each row -
MARGIN = 2: apply function on each column
Let’s create an example dataset:
Age Weight Height SBP
1 32.07784 83.90014 1.693763 128.9139
2 42.16392 63.98754 1.805053 138.3347
3 48.41897 66.17434 1.625191 136.2633
4 47.97991 93.37141 1.621735 133.6035
5 57.89138 70.15537 1.673115 134.0101
6 38.41908 79.18019 1.595290 138.0941Let’s calculate the mean value of each column:
dat_column_mean <- apply(dat, MARGIN = 2, FUN = mean)
dat_column_mean Age Weight Height SBP
39.784181 78.426632 1.729168 133.145297 Hint: It is possibly easiest to think of the “MARGIN” as the dimension you want to keep.
In the above case, we want the mean for each variable, i.e. we want to keep columns and collapse rows.
Purely as an example to understand what apply() does, here is the equivalent procedure using a for-loop. You notice how much more code is needed, and why apply() and similar functions might be very convenient for many different tasks.
Age Weight Height SBP
39.784181 78.426632 1.729168 133.145297 Let’s create a different example dataset, where we record weight at multiple timepoints:
dat2 <- data.frame(ID = seq(8001, 8020),
Weight_week_1 = rnorm(20, mean = 110, sd = 10))
dat2[["Weight_week_3"]] <- dat2[["Weight_week_1"]] + rnorm(20, mean = -2, sd = 1)
dat2[["Weight_week_5"]] <- dat2[["Weight_week_3"]] + rnorm(20, mean = -3, sd = 1.1)
dat2[["Weight_week_7"]] <- dat2[["Weight_week_5"]] + rnorm(20, mean = -1.8, sd = 1.3)
dat2 ID Weight_week_1 Weight_week_3 Weight_week_5 Weight_week_7
1 8001 112.28341 109.96613 107.24360 105.22171
2 8002 118.01189 115.53341 110.92989 110.52237
3 8003 121.80750 120.53422 118.15328 114.94570
4 8004 113.80664 111.89084 108.66007 108.29819
5 8005 108.94540 106.89152 101.83864 101.70218
6 8006 99.18306 96.97893 94.39121 92.87800
7 8007 105.47323 103.23795 100.12770 98.58883
8 8008 113.33714 110.93524 106.85304 104.27920
9 8009 119.97662 117.45126 113.62881 113.16200
10 8010 109.18764 107.59331 104.39319 102.86561
11 8011 118.27388 115.91925 112.08385 111.84434
12 8012 105.00028 104.08398 102.04976 98.49953
13 8013 122.34850 119.69478 116.85126 116.45418
14 8014 111.58446 110.26855 108.28318 107.13491
15 8015 100.31358 99.56235 96.36517 93.39731
16 8016 107.25196 104.25518 101.13239 99.51525
17 8017 106.05919 105.05945 105.05107 102.12757
18 8018 96.61775 95.94047 94.50916 90.50821
19 8019 117.95531 117.13277 114.30635 110.76952
20 8020 105.01579 101.27245 96.89768 96.15095Let’s get the mean weight per week:
apply(dat2[, -1], 2, mean)Weight_week_1 Weight_week_3 Weight_week_5 Weight_week_7
110.6217 108.7101 105.6875 103.9433 Let’s get the mean weight per individual across all weeks:
apply(dat2[, -1], 1, mean) [1] 108.67871 113.74939 118.86017 110.66393 104.84444 95.85780 101.85693
[8] 108.85115 116.05468 106.00994 114.53033 102.40839 118.83718 109.31778
[15] 97.40960 103.03869 104.57432 94.39390 115.04099 99.83422apply() converts 2-dimensional objects to matrices before applying the function. Therefore, if applied on a data.frame with mixed data types, it will be coerced to a character matrix.
This is explained in the apply() documentation under “Details”:
“If X is not an array but an object of a class with a non-null dim value (such as a data frame), apply attempts to coerce it to an array via as.matrix if it is two-dimensional (e.g., a data frame) or via as.array.”
Because of the above, see what happens when you use apply on the iris data.frame which contains 4 numeric variables and one factor:
str(iris)'data.frame': 150 obs. of 5 variables:
$ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
$ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
$ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
$ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
$ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...apply(iris, 2, class)Sepal.Length Sepal.Width Petal.Length Petal.Width Species
"character" "character" "character" "character" "character"
24.2 lapply()
lapply() applies a function on each element of its input and returns a list of the outputs.
Note: The ‘elements’ of a data frame are its columns (remember, a data frame is a list with equal-length elements). The ‘elements’ of a matrix are each cell one by one, by column. Therefore, unlike apply(), lapply() has a very different effect on a data frame and a matrix. lapply() is commonly used to iterate over the columns of a data frame.
lapply() is the only function of the *apply() family that always returns a list.
dat_median <- lapply(dat, median)
dat_median$Age
[1] 41.00496
$Weight
[1] 79.78681
$Height
[1] 1.742986
$SBP
[1] 133.6381To understand what lapply() does, here is the equivalent for-loop:
24.3 sapply()
sapply() is an alias for lapply(), followed by a call to simplify2array().
(Check the source code for sapply() by typing sapply at the console).
dat_median <- sapply(dat, median)
dat_median Age Weight Height SBP
41.004961 79.786811 1.742986 133.638093 dat_summary <- data.frame(Mean = sapply(dat, mean),
SD = sapply(dat, sd))
dat_summary Mean SD
Age 39.784181 8.7788615
Weight 78.426632 9.5932982
Height 1.729168 0.1322217
SBP 133.145297 4.425086624.3.1 Example: Get index of numeric variables
Let’s use sapply() to get an index of numeric columns in dat2:
head(dat2) ID Weight_week_1 Weight_week_3 Weight_week_5 Weight_week_7
1 8001 112.28341 109.96613 107.24360 105.2217
2 8002 118.01189 115.53341 110.92989 110.5224
3 8003 121.80750 120.53422 118.15328 114.9457
4 8004 113.80664 111.89084 108.66007 108.2982
5 8005 108.94540 106.89152 101.83864 101.7022
6 8006 99.18306 96.97893 94.39121 92.8780logical index of numeric columns:
numidl <- sapply(dat2, is.numeric)
numidl ID Weight_week_1 Weight_week_3 Weight_week_5 Weight_week_7
TRUE TRUE TRUE TRUE TRUE integer index of numeric columns:
24.4 Anonymous functions
Anonymous functions are just like regular functions but they are not assigned to an object - i.e. they are not “named”.
They are usually passed as arguments to other functions to be used once, hence no need to assign them.
Anonymous functions are often used with the apply family of functions.
Example of a simple regular function:
squared <- function(x) {
x^2
}Since this is a short function definition, it can also be written in a single line without the curly braces:
squared <- function(x) x^2An anonymous function definition is just like a regular function - minus it is not assigned:
function(x) x^2Since R version 4.1 (May 2021), a compact anonymous function syntax is available, where a single back slash replaces function:
\(x) x^2Let’s use the squared() function within sapply() to square the first four columns of the iris dataset. In these examples, we often wrap functions around head() which prints the first few lines of an object to avoid:
head(dat[, 1:4]) Age Weight Height SBP
1 32.07784 83.90014 1.693763 128.9139
2 42.16392 63.98754 1.805053 138.3347
3 48.41897 66.17434 1.625191 136.2633
4 47.97991 93.37141 1.621735 133.6035
5 57.89138 70.15537 1.673115 134.0101
6 38.41908 79.18019 1.595290 138.0941 Age Weight Height SBP
[1,] 1028.988 7039.233 2.868834 16618.80
[2,] 1777.797 4094.406 3.258218 19136.50
[3,] 2344.397 4379.043 2.641246 18567.68
[4,] 2302.072 8718.220 2.630026 17849.91
[5,] 3351.411 4921.776 2.799313 17958.71
[6,] 1476.026 6269.502 2.544950 19069.98Let’s do the same as above, but this time using an anonymous function:
Age Weight Height SBP
[1,] 1028.988 7039.233 2.868834 16618.80
[2,] 1777.797 4094.406 3.258218 19136.50
[3,] 2344.397 4379.043 2.641246 18567.68
[4,] 2302.072 8718.220 2.630026 17849.91
[5,] 3351.411 4921.776 2.799313 17958.71
[6,] 1476.026 6269.502 2.544950 19069.98The entire anonymous function definition is passed to the FUN argument.
24.5 vapply()
Much less commonly used (possibly underused) than lapply() or sapply(), vapply() allows you to specify what the expected output looks like - for example a numeric vector of length 2, a character vector of length 1.
This can have two advantages:
- It is safer against errors
- It will sometimes be a little faster
You add the argument FUN.VALUE which must be of the correct type and length of the expected result of each iteration.
vapply(dat, median, FUN.VALUE = 0.0) Age Weight Height SBP
41.004961 79.786811 1.742986 133.638093 Here, each iteration returns the median of each column, i.e. a numeric vector of length 1.
Therefore FUN.VALUE can be any numeric scalar.
For example, if we instead returned the range of each column, FUN.VALUE should be a numeric vector of length 2:
Age Weight Height SBP
[1,] 21.53906 51.62685 1.441933 122.3622
[2,] 60.83426 101.09714 1.996182 141.8429If FUN.VALUE does not match the returned value, we get an informative error:
vapply(dat, range, FUN.VALUE = 0.0)Error in vapply(dat, range, FUN.VALUE = 0): values must be length 1,
but FUN(X[[1]]) result is length 2
24.6 tapply()
tapply() is one way (of many) to apply a function on subgroups of data as defined by one or more factors.
Age Weight Height SBP Group
1 32.07784 83.90014 1.693763 128.9139 B
2 42.16392 63.98754 1.805053 138.3347 C
3 48.41897 66.17434 1.625191 136.2633 A
4 47.97991 93.37141 1.621735 133.6035 A
5 57.89138 70.15537 1.673115 134.0101 A
6 38.41908 79.18019 1.595290 138.0941 Cmean_Age_by_Group <- tapply(dat[["Age"]], dat[["Group"]], mean)
mean_Age_by_Group A B C
42.05854 38.86803 38.08844 The for-loop equivalent of the above is:
# Get the group names we want to iterate over
groups <- levels(dat[["Group"]])
# Initialize an empty numeric vector
mean_Age_by_Group <- vector("numeric", length = length(groups))
# Assign names to the initialized vector
names(mean_Age_by_Group) <- groups
# Iterate over the groups and assign the mean Age of each group to the vector
for (i in seq(groups)) {
mean_Age_by_Group[i] <-
mean(dat[["Age"]][dat[["Group"]] == groups[i]])
}
mean_Age_by_Group A B C
42.05854 38.86803 38.08844
24.7 mapply()
The functions we have looked at so far work well when you iterating over elements of a single object.
mapply() allows you to execute a function that accepts two or more inputs, say fn(x, z) using the i-th element of each input, and will return:fn(x[1], z[1]), fn(x[2], z[2]), …, fn(x[n], z[n])
Let’s create a simple function that accepts two numeric arguments, and two vectors length 5 each:
raise <- function(x, power) x^power
x <- 2:6
p <- 6:2Use mapply to raise each x to the corresponding p:
out <- mapply(raise, x, p)
out[1] 64 243 256 125 36This is only for demonstration. In practice, you would use vectorization:
x^p[1] 64 243 256 125 36The equivalent for-loop is:
24.8 *apply()ing on matrices vs. data frames
To consolidate some of what was learned above, let’s focus on the difference between working on a matrix vs. a data frame.
First, let’s create a matrix and a data frame with the same data:
Feature_1 Feature_2 Feature_3 Feature_4 Feature_5
[1,] 21 31 41 51 61
[2,] 22 32 42 52 62
[3,] 23 33 43 53 63
[4,] 24 34 44 54 64
[5,] 25 35 45 55 65
[6,] 26 36 46 56 66
[7,] 27 37 47 57 67
[8,] 28 38 48 58 68
[9,] 29 39 49 59 69
[10,] 30 40 50 60 70adf <- as.data.frame(amat)
adf Feature_1 Feature_2 Feature_3 Feature_4 Feature_5
1 21 31 41 51 61
2 22 32 42 52 62
3 23 33 43 53 63
4 24 34 44 54 64
5 25 35 45 55 65
6 26 36 46 56 66
7 27 37 47 57 67
8 28 38 48 58 68
9 29 39 49 59 69
10 30 40 50 60 70We’ve seen that with apply() we specify the dimension to operate on and it works the same way on both matrices and data frames:
apply(amat, 2, mean)Feature_1 Feature_2 Feature_3 Feature_4 Feature_5
25.5 35.5 45.5 55.5 65.5 apply(adf, 2, mean)Feature_1 Feature_2 Feature_3 Feature_4 Feature_5
25.5 35.5 45.5 55.5 65.5 However, sapply() (and lapply(), vapply()) acts on each element of the object, therefore it is not meaningful to pass a matrix to it:
sapply(amat, mean) [1] 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45
[26] 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70The above returns the mean of each element, i.e. the element itself, which is meaningless.
Since a data frame is a list, and its columns are its elements, it works great for column operations on data frames:
sapply(adf, mean)Feature_1 Feature_2 Feature_3 Feature_4 Feature_5
25.5 35.5 45.5 55.5 65.5 If you want to use sapply() on a matrix, you could iterate over an integer sequence as shown in the previous section:
This is shown to help emphasize the differences between the function and the data structures. In practice, you would use apply() on a matrix.
24.9 Iterating over a sequence instead of an object
With lapply(), sapply() and vapply() there is a very simple trick that may often come in handy:
Instead of iterating over elements of an object, you can iterate over an integer index of whichever elements you want to access
This approach is closer to how we would use an integer sequence in a for loop.
It will be clearer through an example, where we get the mean of each column:
The straightforward use of sapply() to get the mean of every column:
Warning in mean.default(i): argument is not numeric or logical: returning NA Age Weight Height SBP Group
39.784181 78.426632 1.729168 133.145297 NA Just for demonstraion, iterate over integer index of the elements:
Notice that in the above approach you are not passing the object (dat) to lapply(). You therefore need to access it within the anonymous function.
Equivalent to:
for (i in 1:4) {
mean(dat[, i])
}
24.10 replicate()
replicate() is a wrapper around sapply() that is useful when you want to repeat an expression multiple times, for example to perform a simulation study.
This is equivalent to:
24.11 Map()
Map() is a wrapper around mapply() with SIMPLIFY = FALSE, making it more predictable (always returns a list, like lapply()):
Map(function(x, y) x + y, 1:5, 6:10)[[1]]
[1] 7
[[2]]
[1] 9
[[3]]
[1] 11
[[4]]
[1] 13
[[5]]
[1] 15
24.12 Reduce()
Reduce() is a function that iteratively applies a binary function (a function that takes two arguments) to the elements of a vector or list, reducing it to a single value. It uses for loops internally.
Let’s start with a simple example to understand how Reduce() works:
[1] 350The above is equivalent to:
sum(daily_doses)[1] 350In this case, Reduce() gives us the same result as sum(), so it’s not useful. However, it helps us understand what’s happening: Reduce() takes the first two elements (50 + 50 = 100), then adds the third (100 + 50 = 150), then the fourth (150 + 50 = 200), and so on.
Reduce() becomes much more useful when we set accumulate = TRUE, which returns all the intermediate results:
# Track cumulative medication dose across the week
cumulative_doses <- Reduce(`+`, daily_doses, accumulate = TRUE)
cumulative_doses[1] 50 100 150 200 250 300 350Now we can see the cumulative dose after each day, which is clinically relevant for monitoring total drug exposure over time.
Here’s a more complex example where Reduce() is truly useful - calculating drug concentration after multiple doses, accounting for both accumulation and decay between doses:
# Simulate drug concentration after multiple doses
# Each dose adds 100mg, but concentration decays by 30% between doses
doses <- rep(100, 5) # 5 doses of 100mg each
# Function: current concentration + new dose, after 30% decay
accumulate_drug <- function(current, new_dose) {
current * 0.7 + new_dose
}
concentrations <- Reduce(accumulate_drug, doses, accumulate = TRUE)
concentrations[1] 100.00 170.00 219.00 253.30 277.31This shows the concentration after each dose, accounting for the fact that some of the previous dose remains in the system (70% of it) when the next dose is administered.